On January 13, 2026, Google Research unveiled MedGemma 1.5 — the latest evolution of its open-source medical AI family under the Health AI Developer Foundations (HAI-DEF) program.
 Built on the lightweight Gemma architecture, this 4B-parameter multimodal model significantly expands capabilities in high-dimensional medical imaging while delivering substantial gains in text-based clinical reasoning. Released alongside it is MedASR, a specialized open automated speech recognition (ASR) model tuned for medical dictation, slashing transcription errors in healthcare workflows.
Built on the lightweight Gemma architecture, this 4B-parameter multimodal model significantly expands capabilities in high-dimensional medical imaging while delivering substantial gains in text-based clinical reasoning. Released alongside it is MedASR, a specialized open automated speech recognition (ASR) model tuned for medical dictation, slashing transcription errors in healthcare workflows.
These updates arrive as healthcare continues adopting generative AI at roughly twice the rate of the broader economy, driven by the need for reliable tools that handle complex, domain-specific data like 3D scans, pathology slides, electronic health records (EHRs), and spoken clinical notes.
MedGemma 1.5: From 2D to Full 3D Medical Vision + Stronger Text Reasoning
 MedGemma 1.5 builds directly on the original MedGemma collection (released in 2025), which already supported 2D radiology (chest X-rays), dermatology, fundus photography, and histopathology patches.
MedGemma 1.5 builds directly on the original MedGemma collection (released in 2025), which already supported 2D radiology (chest X-rays), dermatology, fundus photography, and histopathology patches.
The 1.5 update introduces native support for:
- High-dimensional 3D imaging: Full volumetric interpretation of CT scans and MRI series, allowing developers to feed multiple slices together with prompts for holistic analysis (e.g., detecting correlations across lung nodules or brain lesions that single-slice views might miss).
- Whole-slide histopathology imaging (WSI): Simultaneous processing of multiple patches from gigapixel digital slides, enabling context-aware pathology report generation or tumor grading.
- Longitudinal radiology: Improved handling of time-series chest X-rays (e.g., comparing prior and current studies for progression tracking).
- Anatomical localization & structured extraction: Better bounding-box style localization in images and structured data pulling from unstructured lab reports.
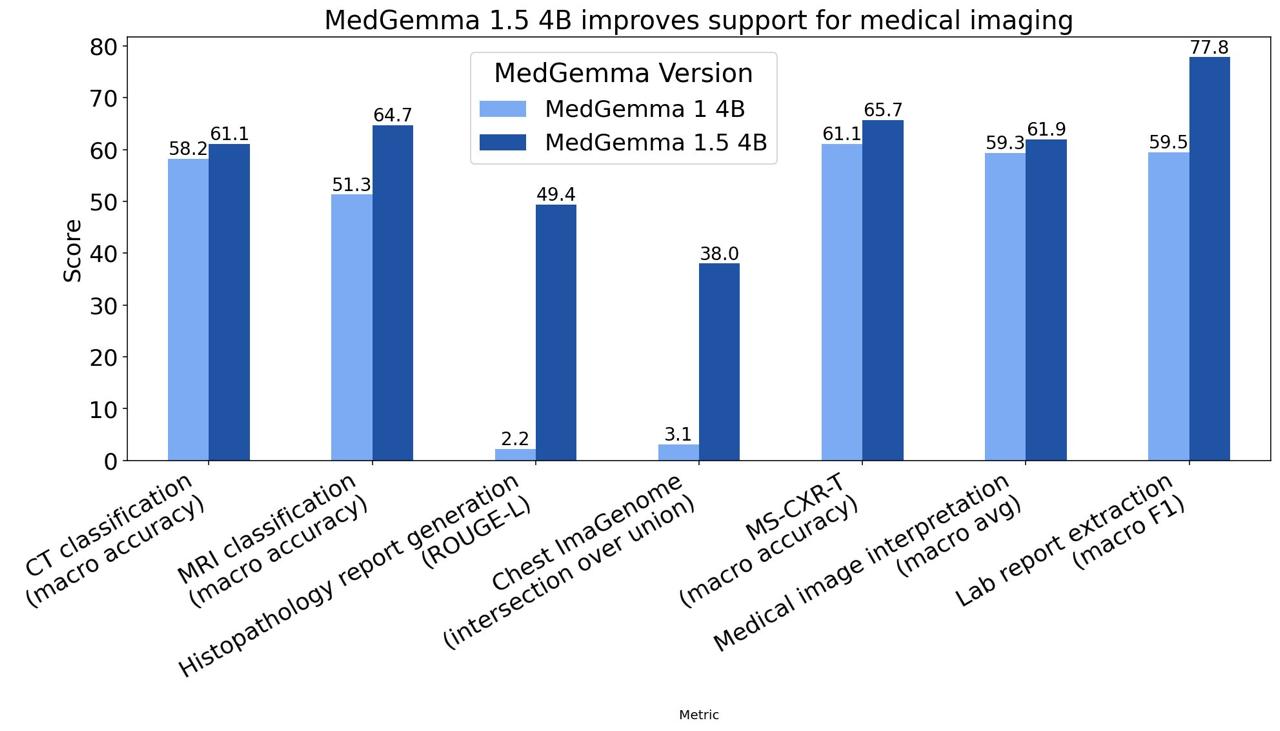
 Internal benchmarks show impressive lifts over MedGemma 1 (4B variant):
Internal benchmarks show impressive lifts over MedGemma 1 (4B variant):
- Disease-related findings classification on CT volumes: +3% absolute accuracy (61% vs. 58%).
- Disease-related findings on MRI: +14% absolute (65% vs. 51%).
- Histopathology report fidelity (ROUGE-L on single-slide cases): massive jump from 0.02 to 0.49 — matching performance of specialized models like PolyPath.
- Chest X-ray anatomical localization (Chest ImaGenome IoU): from 3% to 38% (+35 percentage points).
- Longitudinal chest X-ray comparison (MS-CXR-T macro-accuracy): +5% (66% vs. 61%).
- Average accuracy across internal single-image benchmarks (radiology, derm, histo, ophtho): +3% (62% vs. 59%).
- Lab report information extraction (F1): +18% (78% vs. 60%).
On the text side, MedGemma 1.5 strengthens core medical reasoning:
- MedQA (USMLE-style multiple-choice): +5% (69% vs. 64%).
- EHRQA (question answering over electronic health records): +22% (90% vs. 68%).
The 4B size remains compute-efficient — suitable for offline/local deployment on modest hardware — while developers can still leverage the larger MedGemma 1 27B for heavy text-only tasks. Full DICOM support is now integrated into Google Cloud Vertex AI deployments, simplifying hospital workflow integration.
MedASR: Medical-Grade Speech-to-Text That Actually Understands Doctors
 Complementing the vision upgrades is **MedASR**, a Conformer-based open ASR model (≈105M parameters) fine-tuned on diverse de-identified medical speech data. It targets dictation-heavy scenarios: radiology reports, operative notes, patient consultations, and physician dictations.
Complementing the vision upgrades is **MedASR**, a Conformer-based open ASR model (≈105M parameters) fine-tuned on diverse de-identified medical speech data. It targets dictation-heavy scenarios: radiology reports, operative notes, patient consultations, and physician dictations.
Compared to general-purpose models like OpenAI's Whisper large-v3:
- Chest X-ray dictation word error rate (WER): 5.2% vs. 12.5% → 58% fewer errors.
- Broad internal medical dictation benchmark (multiple specialties, accents, noisy environments): 5.2% vs. 28.2% WER → 82% fewer errors.
This dramatic reduction in hallucinations around drug names, anatomical terms, abbreviations, and proper nouns makes MedASR viable for production transcription pipelines. Outputs can feed directly into MedGemma for end-to-end workflows: dictate a finding → transcribe accurately → query the image model for interpretation or report generation.
Both models are released under permissive licenses for research and commercial use, available on Hugging Face (MedGemma 1.5 4B, MedASR) and Vertex AI. Google also launched tutorial notebooks for CT volume processing, whole-slide histopathology, and integration examples, plus a $100,000 Kaggle challenge to spur community fine-tuning and applications.
Also read:
- The AI Coding Surge: Redefining Productivity, Organizations, and the Future of Work
- The Simplest AI Prompt Hack: Repeat Yourself for Smarter Responses
- Crowded Skies: Starlink's Orbital Shift to Ease Satellite Congestion
Why This Matters: Democratizing Advanced Medical AI
 MedGemma 1.5 marks the first open multimodal LLM capable of natively handling 3D CT/MRI volumes and gigapixel pathology slides without proprietary restrictions — a capability previously locked behind closed systems or massive compute budgets.
MedGemma 1.5 marks the first open multimodal LLM capable of natively handling 3D CT/MRI volumes and gigapixel pathology slides without proprietary restrictions — a capability previously locked behind closed systems or massive compute budgets.
Paired with MedASR's near-clinical transcription accuracy, the duo enables seamless "see-listen-reason" pipelines that mirror real clinician cognition.
Early real-world traction already exists: Malaysia's Ministry of Health uses a MedGemma-powered askCPG tool for navigating 150+ clinical guidelines, while Taiwan's National Health Insurance analyzed 30,000+ pathology reports for lung cancer surgical planning.
Developers are encouraged to validate, fine-tune, and rigorously test adaptations before clinical deployment — as with all foundation models in healthcare.
But with weights, code, and documentation fully open, MedGemma 1.5 + MedASR lower the barrier dramatically for building reliable, multimodal medical AI tools at scale.
Explore the release here:
https://research.google/blog/next-generation-medical-image-interpretation-with-medgemma-15-and-medical-speech-to-text-with-medasr/
Model cards, weights, and notebooks await on Hugging Face and Google Cloud. The next wave of healthcare AI just got a powerful, accessible upgrade.






